Nonparametric Regression
Given the usual conditional expectation function
we can estimate as a traditional linear model or nonparametrically. One approach to nonparametric estimation is to use a binned means estimator. This takes the average for some values of close to (assuming is continuous). In other words, take the average for all values where for some small . Here is the bandwidth.
Binned means estimator
This gives us a step-function because the indicator function makes the weights discontinuous. Alternatively, we can replace the indicator function with a kernel function:
Kernel regression estimator
Some commonly used kernels:
Gaussian Kernel:
Epanechnikov Kernel:
Local Linear approximation
Let’s start with simulating some data using the simstudy packge:
library(tidyverse)
## simulate data using simstudy package
library(simstudy)
set.seed(234)
df <- genData(100, defData(varname = "x", formula = "20;60", dist = 'uniform'))
theta1 = c(0.1, 0.8, 0.6, 0.4, 0.6, 0.9, 0.9)
knots <- c(0.25, 0.5, 0.75)
df <- genSpline(dt = df, newvar = "y",
predictor = "x",
theta = theta1,
knots = knots,
degree = 3,
newrange = "90;160",
noise.var = 64)Here is a scatterplot with dashed vertical lines showing the knots used to generate the data (quantiles).
## scatter plot of the data
ggplot(data = df, aes(x = x, y = y)) +
geom_point() +
geom_vline(xintercept = quantile(df$x, knots), linetype = 'dashed') +
theme_bw()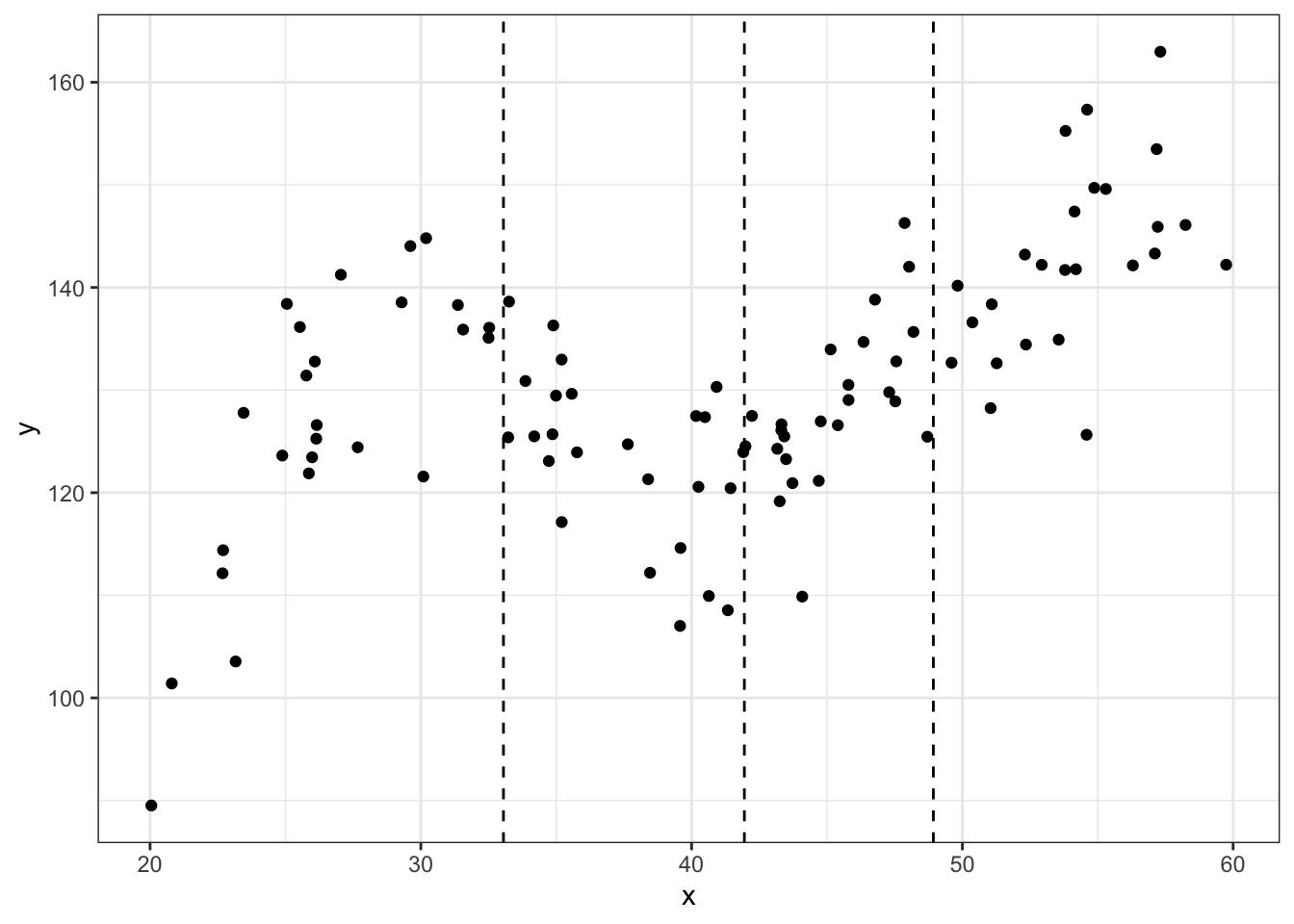
#using locpoly
library(KernSmooth)## KernSmooth 2.23 loaded
## Copyright M. P. Wand 1997-2009m_ll <- data.frame(locpoly(df$x, df$y, degree = 1, bandwidth = 3))
# degree = 1 for locally linear
ggplot(data = df, aes(x = x, y = y)) +
geom_point() +
geom_vline(xintercept = quantile(df$x, knots), linetype = 'dashed') +
geom_line(data = m_ll,aes(x = x, y = y),color = "blue") +
theme_bw()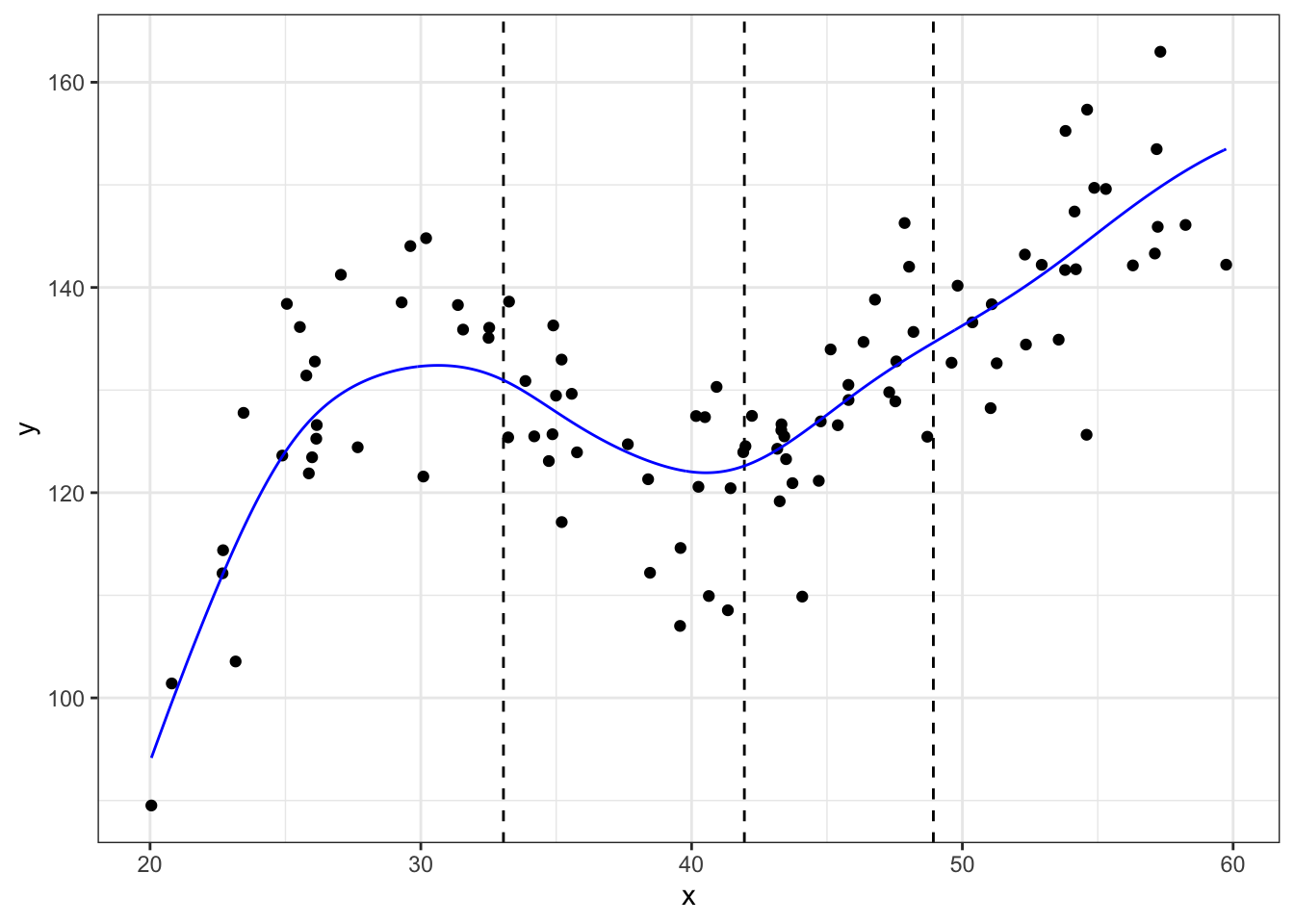
tricube kernal
kernel.function <- function(u){
tmp <- rep(NA,length(u))
for(i in 1:length(u)){
if(abs(u[i])<1){
k <- (1 - abs(u[i])^3)^3
}else if(abs(u[i])>=1){
k <- 0
}
tmp[i] <- k
}
return(tmp)
}Decide on evaluation points . Our data is (roughly) from 20 to 60 so let’s use that to define our evaluation points and we will use increments of 20.
# evaluation points
x0 <- seq(25,55,10)
# bandwidth
h <- 10We have 3 evaluation points, let’s do a loop to calculate the scaled distance from each value of to .
# empty container
distances <- matrix(NA, nrow = length(df$x), ncol = 4)
colnames(distances) <- x0
for(i in 1:length(x0)){
# calc distance and put in column i
distances[,i] <- df$x - x0[i]
# divide by sum of bandwidth (this does scaling)
# want scaled so can use as weights
#distances[,i] <- distances[,i]/sum(distances[,i])
}Now apply our kernel function to estimate the weights
distances <- data.frame(distances)
weights <- map_df(distances, kernel.function)Estimate predicted values at the evaluation points using our weights
# eval point 20
W_25 <- diag(weights$X25)
X <- as.matrix(cbind(rep(1,100), df$x)) #create X matrix with intercept
Y <- as.matrix(df$y)
B_WLS_25 <- solve(t(X)%*%W_25%*%X)%*%(t(X)%*%W_25%*%Y)
# eval point 40
W_45 <- diag(weights$X45)
B_WLS_45 <- solve(t(X)%*%W_45%*%X)%*%(t(X)%*%W_45%*%Y)
# eval point 60
W_55 <- diag(weights$X55)
B_WLS_55 <- solve(t(X)%*%W_55%*%X)%*%(t(X)%*%W_55%*%Y)
# predicted values
yhat <- data.frame(y25 = X%*%B_WLS_25,
y45 = X%*%B_WLS_45,
y55 = X%*%B_WLS_55)
yhat <- pivot_longer(yhat, names_to = "group", names_prefix = "y",
values_to="yhat",cols = c(1:3))
df_big <- bind_cols(bind_rows(df,df,df),yhat)
#first eval point
ggplot(data = df_big %>% filter(group==25), aes(x = x, y = y)) +
geom_point() +
geom_point(aes(x = x, y = yhat),color="red") +
theme_bw()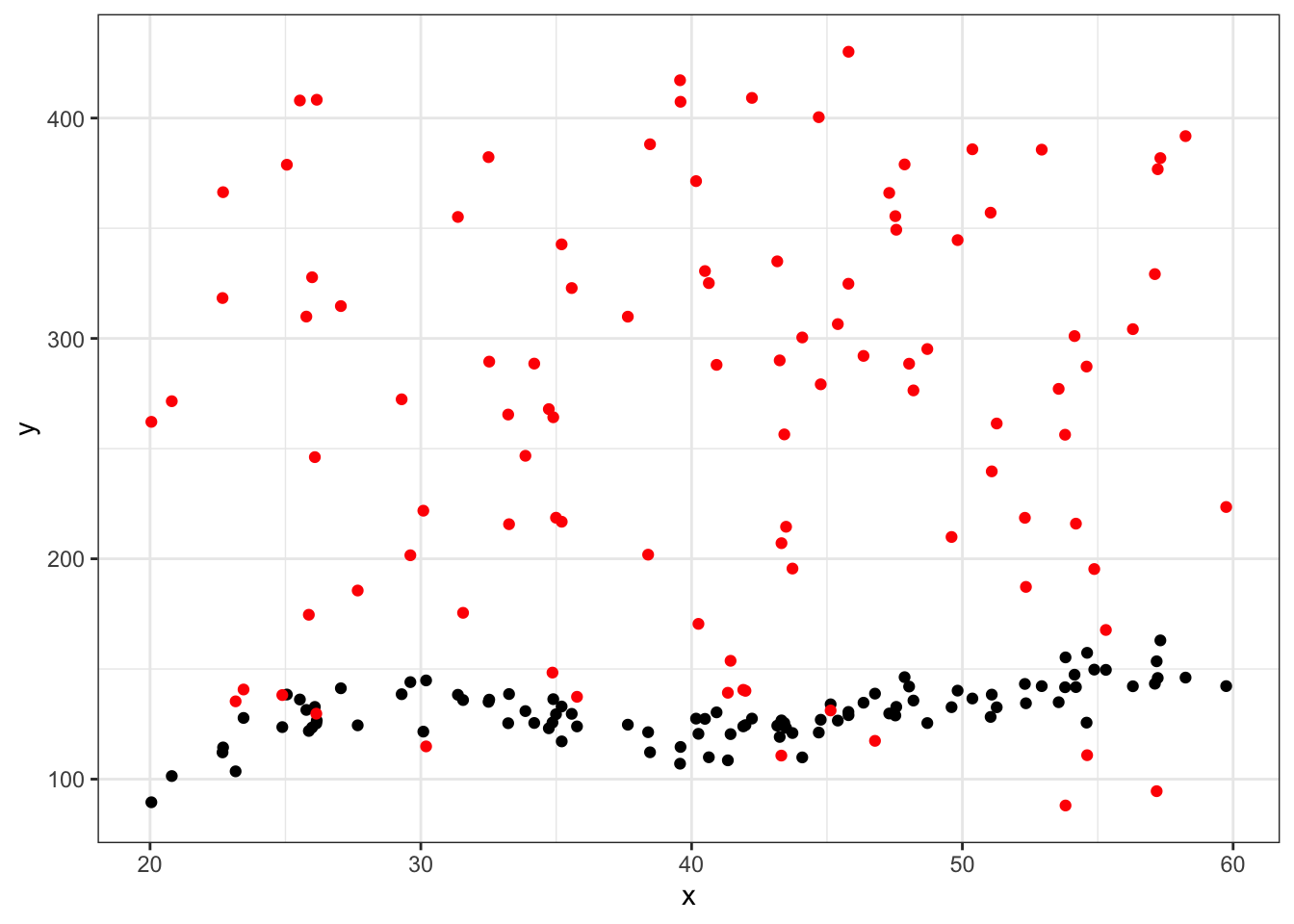
ggplot(data = df_big %>% filter(group==45), aes(x = x, y = y)) +
geom_point() +
geom_point(aes(x = x, y = yhat), color="green") +
theme_bw()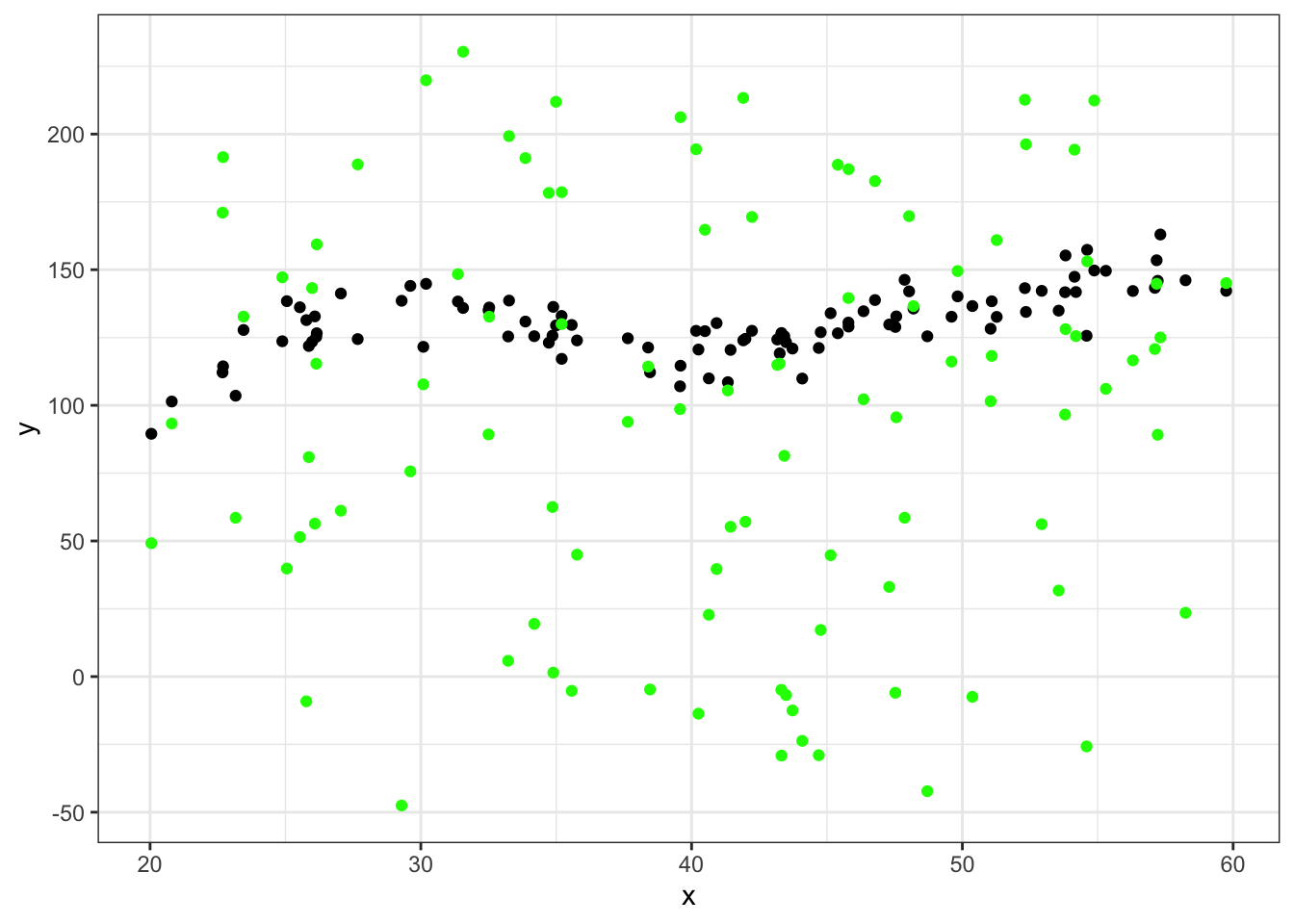
ggplot(data = df_big %>% filter(group==60), aes(x = x, y = y)) +
geom_point() +
geom_point(aes(x = x, y = yhat), color="blue") +
theme_bw()
ggplot(data = df_big, aes(x = x, y = y)) +
geom_point() +
geom_point(aes(x = x, y = yhat, color=group)) +
theme_bw()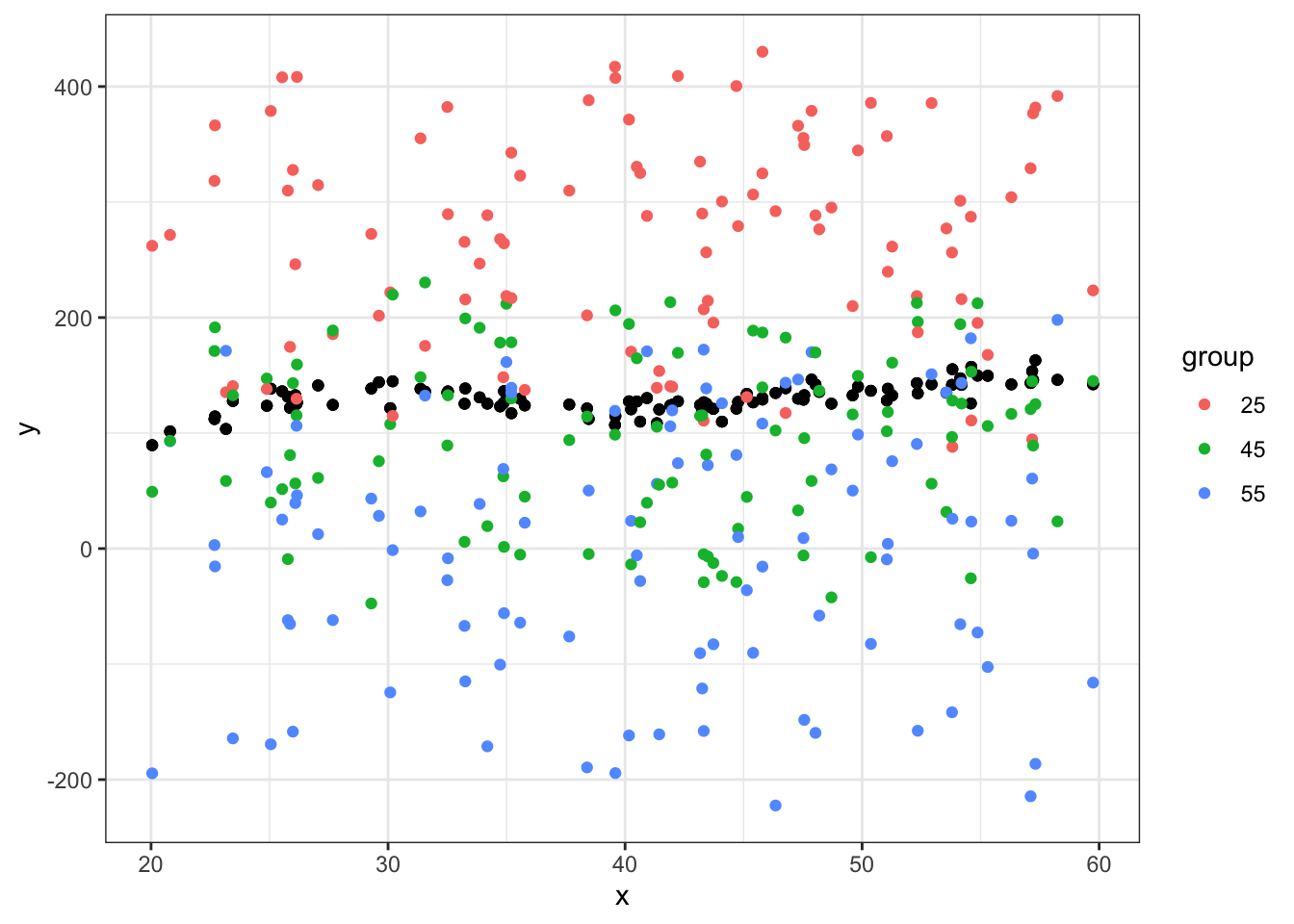
Switching gears – let’s do a quick example on how to demean variables using dplyr. We will use the built-in mtcars dataset.
First, let’s demean only one column, the mpg column and make a new variable using mutate():
df <- mtcars %>%
mutate(mpg_demean = mpg - mean(mpg))You have to assign this to an object, in this case I’ve stored it as an entirely new data.frame df. I can now use this new variable:
head(df$mpg_demean)## [1] 0.909375 0.909375 2.709375 1.309375 -1.390625 -1.990625Here, the demeaned variable is calculated but not stored because I have not assigned this operation to anything. If I try to call the variable mpg_demean later I will get an error because it does not exist.
mtcars %>%
mutate(mpg_demean2 = mpg - mean(mpg)) %>%
head()## mpg cyl disp hp drat wt qsec vs am gear carb mpg_demean2
## 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 0.909375
## 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 0.909375
## 3 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 2.709375
## 4 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 1.309375
## 5 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 -1.390625
## 6 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 -1.990625head(df$mpg_demean2)## NULLIf I want to de-mean multiple variables at once use mutate_each. Here I am selecting two columns, vs and am, then using mutate_all to demean them both then I’m joining them back to the original data.
df <- df %>%
select(vs, am) %>%
mutate_all(list(~. - mean(.))) %>%
bind_cols(df,.)## New names:
## * vs -> vs...8
## * am -> am...9
## * vs -> vs...13
## * am -> am...14head(df)## mpg cyl disp hp drat wt qsec vs...8 am...9 gear carb mpg_demean vs...13
## 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 0.909375 -0.4375
## 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 0.909375 -0.4375
## 3 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 2.709375 0.5625
## 4 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 1.309375 0.5625
## 5 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 -1.390625 -0.4375
## 6 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 -1.990625 0.5625
## am...14
## 1 0.59375
## 2 0.59375
## 3 0.59375
## 4 -0.40625
## 5 -0.40625
## 6 -0.40625Using base R you could do something like this:
df$wt_demean <- df$wt - mean(df$wt)
head(df)## mpg cyl disp hp drat wt qsec vs...8 am...9 gear carb mpg_demean vs...13
## 1 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 0.909375 -0.4375
## 2 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 0.909375 -0.4375
## 3 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 2.709375 0.5625
## 4 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 1.309375 0.5625
## 5 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 -1.390625 -0.4375
## 6 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 -1.990625 0.5625
## am...14 wt_demean
## 1 0.59375 -0.59725
## 2 0.59375 -0.34225
## 3 0.59375 -0.89725
## 4 -0.40625 -0.00225
## 5 -0.40625 0.22275
## 6 -0.40625 0.24275If I want to demean by groups. Here I will use the cylinder variable to group with.
df <- df %>%
group_by(cyl) %>%
mutate(mpg_mean_cyl = mpg - mean(mpg))
head(df[,c("mpg","cyl","mpg_mean_cyl")])## # A tibble: 6 x 3
## # Groups: cyl [3]
## mpg cyl mpg_mean_cyl
## <dbl> <dbl> <dbl>
## 1 21 6 1.26
## 2 21 6 1.26
## 3 22.8 4 -3.86
## 4 21.4 6 1.66
## 5 18.7 8 3.6
## 6 18.1 6 -1.64Here is an example of some code that runs but may not be doing what we want. What are some differences in this code?
df %>%
group_by(cyl) %>%
mutate(new_mpg = mean(mpg, na.rm=TRUE)) %>%
select(mpg,new_mpg)## Adding missing grouping variables: `cyl`## # A tibble: 32 x 3
## # Groups: cyl [3]
## cyl mpg new_mpg
## <dbl> <dbl> <dbl>
## 1 6 21 19.7
## 2 6 21 19.7
## 3 4 22.8 26.7
## 4 6 21.4 19.7
## 5 8 18.7 15.1
## 6 6 18.1 19.7
## 7 8 14.3 15.1
## 8 4 24.4 26.7
## 9 4 22.8 26.7
## 10 6 19.2 19.7
## # … with 22 more rowsF-test
We use the F-test to look at multiple hypotheses at once.
fit <- lm(mpg ~ cyl + vs + am + gear + carb, data = mtcars)
summary(fit)##
## Call:
## lm(formula = mpg ~ cyl + vs + am + gear + carb, data = mtcars)
##
## Residuals:
## Min 1Q Median 3Q Max
## -5.5403 -1.1582 0.2528 1.2787 5.5597
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 25.1591 7.7570 3.243 0.00323 **
## cyl -1.2239 0.7510 -1.630 0.11521
## vs 0.8784 1.9957 0.440 0.66347
## am 3.5989 1.8694 1.925 0.06522 .
## gear 1.2516 1.4730 0.850 0.40323
## carb -1.4071 0.5368 -2.621 0.01444 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.808 on 26 degrees of freedom
## Multiple R-squared: 0.8179, Adjusted R-squared: 0.7829
## F-statistic: 23.36 on 5 and 26 DF, p-value: 7.418e-09We could do an F-test on the following hypothesis:
This is different than testing the individual hypotheses:
The F-test will compare the two models: 1) model with cyl and vs and 2) model without cyl and vs.
Where is the residual sum of squares for the full model. is the residual sum of squares for the null model (restricted). is how many coefficients are omitted.
full <- lm(mpg ~ cyl + vs + am + gear + carb, data = mtcars)
restrict <- lm(mpg ~ am + gear + carb, data = mtcars)
n <- nrow(mtcars)
k <- 5
q <- 2
SSR_unres <- sum((summary(full)$residuals)^2)
SSR_res <- sum((summary(restrict)$residuals)^2)
SSR_unres## [1] 205.0521SSR_res## [1] 265.9296F <- ((SSR_res - SSR_unres)/q)/(SSR_unres/(n-k-1))
F## [1] 3.8595461 - pf(F,df1 = 2, df2 = n-k-1)## [1] 0.03406177Using linearHypothesis() function from the car package:
library(car)## The following object is masked from 'package:dplyr':
##
## recodelinearHypothesis(full, c("cyl = 0", "vs = 0"))## Linear hypothesis test
##
## Hypothesis:
## cyl = 0
## vs = 0
##
## Model 1: restricted model
## Model 2: mpg ~ cyl + vs + am + gear + carb
##
## Res.Df RSS Df Sum of Sq F Pr(>F)
## 1 28 265.93
## 2 26 205.05 2 60.878 3.8595 0.03406 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Given the p-value we can reject the null hypothesis. This suggests that while individually, the coefficients on cyl and vs were not statistically significant at the 95% level, including them both improves model fit because we reject the null that they are jointly equal to 0.